Romero Lab participates in the Wisconsin Science Festival!
Sameer and Emily's paper on ML from directed evolution data is published in PLoS Comp Biology!
D’Costa S, Hinds EC, Freschlin CR, Song H, Romero PA. Inferring protein fitness landscapes from laboratory evolution experiments. PLoS Comp Biol (2023).
Pete's paper engineering ACE2 as a protein therapeutic is published Protein Science
Heinzelman P, Romero PA. Directed evolution of angiotensin-converting enzyme 2 (ACE2) peptidase activity profiles for therapeutic applications. Protein Science (2023) e4597.
Welcome to new PhD students Nathaniel Blalock and Andres Lira!
Nathaniel is a ChemE grand student coming from Univ Tennessee and and Andres is a Biophysics grad student coming from UC Davis.
Leland's paper developing molecular SNP detection methods is published in Cell Reports Methods!
Hyman LB, Christopher CR, Romero PA. Competitive SNP-LAMP probes for rapid and robust single-nucleotide polymorphism detection. Cell Reports Methods (2022) 2, 100242.
Chase and Sarah's review on ML-guided protein engineering is published in Curr Opin Biotech!
Freschlin CR, Fahlberg SA, Romero PA. Machine learning to navigate fitness landscapes for protein engineering. Curr Opin Biotechnol (2022) 75.
Farewell to Leland, Hridindu, and Jonathan!
The first wave of Romero Lab PhD students. It’s been a fun ride and best of luck with the new positions!
Congrats to Jonathan for an awesome PhD defense!
Congrats to Hridindu for successfully defending his PhD thesis!
Congrats to Hridindu for publishing his caspase deep mutational scanning work in Cell Death Discovery
Roychowdhury H, Romero PA. Microfluidic deep mutational scanning of the human executioner caspases reveals differences in structure and regulation. Cell Death Discov (2022) 8.
https://www.nature.com/articles/s41420-021-00799-0
Welcome to our new graduate students Meg Taylor and Nate Novy!
Our deep learning collaboration with Tony Gitter is published in PNAS!
Gelman S, Fahlberg SA, Heinzelman P, Romero PA*, Gitter A*. Neural networks to learn protein sequence-function relationships from deep mutational scanning data. Proc Natl Acad Sci USA (2021) 118, e2104878118.
https://www.pnas.org/content/118/48/e2104878118
Jonathan's work engineering acyl-ACP reductases is published in Nature Communications!
Greenhalgh JC, Fahlberg SA, Pfleger BF, Romero PA. Machine learning-guided acyl-ACP reductase engineering for improved in vivo fatty alcohol production. Nature Comm (2021) 5825.
https://www.nature.com/articles/s41467-021-25831-w
We've received funding through the WARF Accelerator program to commercialize Leland's microfluidic single-cell analysis methods
News story from Biochemistry: https://biochem.wisc.edu/news/2021/news-phil-romero-warf-accelerator-award-2021-08-03
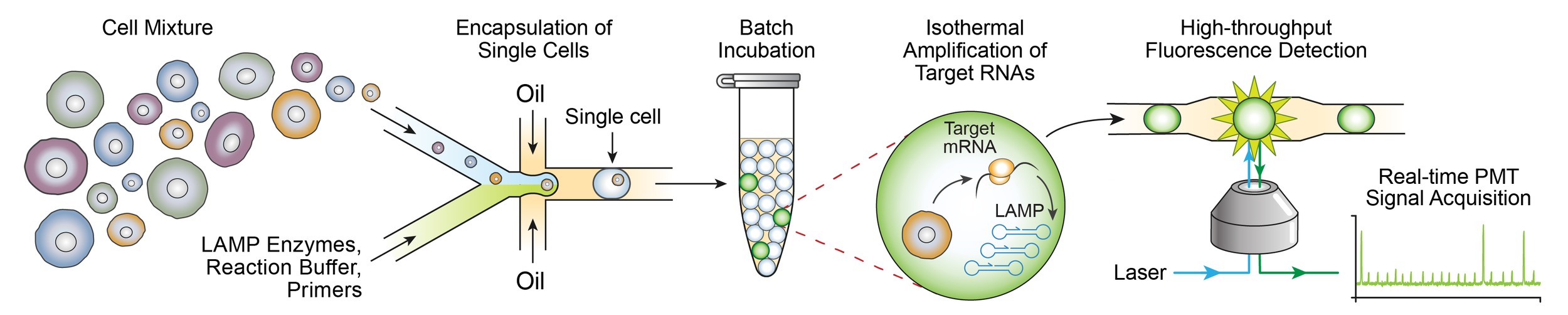
Leland's work on single-cell gene expression profiling is published in Nucleic Acids Research!
Hyman LB, Christopher CR, Romero PA. Single-cell nucleic acid profiling in droplets (SNAPD) enables high-throughput analysis of heterogeneous cell populations. Nucleic Acids Research (2021) https://academic.oup.com/nar/advance-article/doi/10.1093/nar/gkab577/6316829
Our collaborative project to produce carbon-negative fuels and chemicals led by Brian Pfleger was funded by ARPA-E!
Pete's paper mapping ACE2 interactions with SARS-CoV-2 is published in PLoS One!
Heinzelman P, Romero PA. Discovery of human ACE2 variants with altered recognition by the SARS-CoV-2 spike protein. PLoS ONE (2021)
https://journals.plos.org/plosone/article?id=10.1371%2Fjournal.pone.0251585
Jacob and Ben's recent patent application was covered by the Academic Times!
Congratulations to our undergraduates Sarah, Evelyn, and Ankit for receiving summer research fellowships!
Sarah and Evelyn both received the Hilldale Fellowship and the Biochemistry Undergraduate Summer Research Award. Ankit received the Sophomore Honors Summer Apprenticeship. Looking forward to a fun summer!